DNA Molecule
For 3-D Structure of DNA using Jsmol Click here
Deoxyribonucleic acid (DNA) is the primary chemical component of chromosomes and is the material of which genes are made. It is sometimes called the "molecule of heredity," because parents transmit copied portions of their own DNA to offspring during reproduction, and because they propagate their traits by doing so.
In bacteria and other simple or prokaryotic cell organisms, DNA is distributed more or less throughout the cell. In the complex or eukaryotic cells that make up plants, animals and in other multi-celled organisms, most of the DNA resides in the cell nucleus. The energy-generating organelles known as chloroplasts and mitochondria also carry DNA, as do many viruses.
Overview of molecular structure
Although sometimes called "the molecule of heredity," pieces of DNA as people typically think of them are not single molecules. Rather, they are pairs of molecules, which entwine like vines to form a double helix (top half of the illustration at the right).
Each vine-like molecule is a strand of DNA: a chemically linked chain of nucleotides, each of which consists of a sugar, a phosphate and one of four kinds of aromatic "bases". Because DNA strands are composed of these nucleotide subunits, they are polymers.
The diversity of the bases means that there are four kinds of nucleotides, which are commonly referred to by the identity of their bases. These are adenine (A), thymine (T), cytosine (C), and guanine (G).
In a DNA double helix, two polynucleotide strands come together through complementary pairing of the bases, which occurs by hydrogen bonding. Each base forms hydrogen bonds readily to only one other -- A to T and C to G -- so that the identity of the base on one strand dictates what base must face it on the opposing strand. Thus the entire nucleotide sequence of each strand is complementary to that of the other, and when separated, each may act as a template with which to replicate the other (middle and lower half of the illustration at the right).
Because pairing causes the nucleotide bases to face the helical axis, the sugar and phosphate groups of the nucleotides run along the outside, and the two chains they form are sometimes called the "backbones" of the helix. In fact, it is chemical bonds between the phosphates and the sugars that link one nucleotide to the next in the DNA strand.
The role of the sequence
Within a gene, the sequence of nucleotides along a DNA strand defines a protein, which an organism is liable to manufacture or "express" at one or several points in its life using the information of the sequence. The relationship between the nucleotide sequence and the amino-acid sequence of the protein is determined by simple cellular rules of translation, known collectively as the genetic code. Reading along the "protein-coding" sequence of a gene, each successive sequence of three nucleotides (called a codon) specifies or "encodes" one amino acid.
In many species of organism, only a small fraction of the total sequence of the genome appears to encode protein. The function of the rest is a matter of speculation. It is known that certain nucleotide sequences specify affinity for DNA binding proteins, which play a wide variety of vital roles, in particular through control of replication and transcription. These sequences are frequently called regulatory sequences, and researchers assume that so far they have identified only a tiny fraction of the total that exist. "Junk DNA" represents sequences that do not yet appear to contain genes or to have a function.
Sequence also determines a DNA segment's susceptibility to cleavage by restriction enzymes, the quintessential tools of genetic engineering. The position of cleavage sites throughout an individual's genome determines one kind of an individual's "DNA fingerprint".
DNA replication
The hydrogen bonds between the strands of the double helix are weak enough that they can be easily separated by enzymes. Enzymes known as helicases unwind the strands to facilitate the advance of sequence-reading enzymes such as DNA polymerase. The unwinding requires that helicases chemically cleave the phosphate backbone of one of the strands so that it can swivel around the other. The stands can also be separated by gentle heating, as used in PCR, provided they have fewer than about 10,000 base pairs (10 kilobase pairs, or 10 kbp). The intertwining of the DNA strands makes long segments difficult to separate.
When the ends of a piece of double-helical DNA are joined so that it forms a circle, as in plasmid DNA, the strands are topologically knotted. This means they cannot be separated by gentle heating or by any process that does not involve breaking a strand. The task of unknotting topologically linked strands of DNA falls to enzymes known as topoisomerases. Some of these enzymes unknot circular DNA by cleaving two strands so that another double-stranded segment can pass through. Unknotting is required for the replication of circular DNA as well as for various types of recombination in linear DNA.
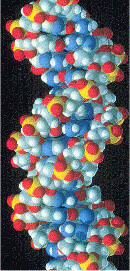
Space-filling model of a section of DNA molecule
The DNA helix can assume one of three slightly different geometries, of which the "B" form described by James D. Watson and Francis Crick is believed to predominate in cells. It is 2 nanometers wide and extends 3.4 nanometers per 10 bp of sequence. This is also the approximate length of sequence in which the helix makes one complete turn about its axis. This frequency of twist (known as the helical pitch) depends largely on stacking forces that each base exerts on its neighbors in the chain.
The narrow breadth of the double helix makes it impossible to detect by conventional electron microscopy, except by heavy staining. At the same time, the DNA found in many cells can be macroscopic in length -- approximately 5 centimeters long for strands in a human chromosome. Consequently, cells must compact or "package" DNA to carry it within them. This is one of the functions of the chromosomes, which contain spool-like proteins known as histones, around which DNA winds.
The B form of the DNA helix twists 360° per 10.6 bp in the absence of strain. But many molecular biological processes can induce strain. A DNA segment with excess or insufficient helical twisting is referred to, respectively, as positively or negatively "supercoiled". DNA in vivo is typically negatively supercoiled, which facilitates the unwinding of the double-helix required for RNA transcription.The two other known double-helical forms of DNA, called A and Z, differ modestly in their geometry and dimensions. The A form appears likely to occur only in dehydrated samples of DNA, such those used in crystallography experiments, and possibly in hybrid pairings of DNA and RNA strands. Segments of DNA that cells have methylated for regulatory purposes may adopt the Z geometry, in which the strands turn about the helical axis like a mirror image of the B form.
DNA sequence reading
The asymmetric shape and linkage of nucleotides means that a DNA strand always has a discernable orientation or directionality. Because of this directionality, close inspection of a double helix reveals that, although the nucleotides along one strand are heading one way (e.g. the "ascending strand") the others are heading the other (e.g. the "descending strand"). This arrangement of the strands is called antiparallel.
For reasons of chemical nomenclature, people who work with DNA refer to the asymmetric termini of each strand as the 5' and 3' ends (pronounced "five prime" and "three prime"). DNA workers and enzymes alike always read nucleotide sequences in the "5' to 3' direction". In a vertically oriented double helix, the 3' strand is said to be ascending while the 5' strand is said to be descending.
As a result of their antiparallel arrangement and the sequence-reading preferences of enzymes, even if both strands carried identical instead of complementary sequences, cells could properly translate only one of them. The other strand a cell can only read backwards. Molecular biologists call a sequence "sense" if it is translated or translatable, and they call its complement "antisense". It follows then, somewhat paradoxically, that the template for transcription is the antisensestrand. The resulting transcript is an RNA replica of the sense strand and is itself sense.
Some viruses blur the distinction between sense and antisense, because certain sequences of their genomes do double duty, encoding one protein when read 5' to 3' along one strand, and a second protein when read in the opposite direction along the other strand. As a result, the genomes of these viruses are unusually compact for the number of genes they contain, which biologists view as an adaptation. Topologists like to note that the juxtaposition of the 3' end of one DNA strand beside the 5' end of the other at both termini of a double-helical segment makes the arrangement a "crab canon".
Single-stranded DNA (ssDNA) and repair of mutations
In some viruses DNA appears in a non-helical, single-stranded form. Because many of the DNA repair mechanisms of cells work only on paired bases, viruses that carry single-stranded DNA genomes mutate more frequently than they would otherwise. As a result, such species may adapt more rapidly to avoid extinction. The result would not be so favorable in more complicated and more slowly replicating organisms, however, which may explain why only viruses carry single-stranded DNA. These viruses presumably also benefit from the lower cost of replicating one strand versus two.
The discovery of DNA and the double helix
Working in the 19th century, biochemists initially isolated DNA and RNA (mixed together) from cell nuclei. They were relatively quick to appreciate the polymeric nature of their "nucleic acid" isolates, but realized only later that nucleotides were of two types--one containing ribose and the other deoxyribose. It was this subsequent discovery that led to the identification and naming of DNA as a substance distinct from RNA.
Friederich Miescher (1844-1895) discovered a substance he called "nuclein" in 1869. Somewhat later he isolated a pure sample of the material now known as DNA from the sperm of salmon, and in 1889 his pupil, Richard Altmann, named it "nucleic acid". This substance was found to exist only in the chromosomes. Max Delbrück, Nikolai V. Timofeeff-Ressovsky, and Karl G. Zimmer published results in 1935 suggesting that chromosomes are very large molecules the structure of which can be changed by treatment with X-rays, and that by so changing their structure it was possible to change the heritable characteristics governed by those chromosomes. (Delbrück and Salvador Luria were awarded the Nobel Prize in 1969 for their work on the genetic structure of viruses.) In 1943, Oswald Theodore Avery discovered that traits proper to the "smooth" form of the Pneumococcus could be transferred to the "rough" form of the same bacteria merely by making the killed "smooth" (S) form available to the live "rough" (R) form. Quite unexpectedly, the living R Pneumococcus bacteria were transformed into a new strain of the S form, and the transferred S characteristics turned out to be heritable.
In 1944, the renowned physicist, Erwin Schrödinger, published a brief book entitled What is Life?, in which he maintained that chromosomes contained what he called the "hereditary code-script" of life. He added: "But the term code-script is, of course, too narrow. The chromosome structures are at the same time instrumental in bringing about the development they foreshadow. They are law-code and executive power -- or, to use another simile, they are architect's plan and builder's craft -- in one." He conceived of these dual functional elements as being woven into the molecular structure of chromosomes. By understanding the exact molecular structure of the chromosomes one could hope to understand both the "architect's plan" and also how that plan was carried out through the "builder's craft." Francis Crick, James Watson, Maurice Wilkins, Seymour Benzer, et al., took up the physicist's challenge to work out the structure of the chromosomes and the question of how the segments of the chromosomes that were conceived to relate to specific traits could possibly do their jobs.
Just how the presence of specific features in the molecular structure of chromosomes could produce traits and behaviors in living organisms was unimaginable at the time. Because chemical dissection of DNA samples always yielded the same four nucleotides, the chemical composition of DNA appeared simple, perhaps even uniform. Organisms, on the other hand, are fantastically complex individually and widely diverse collectively. Geneticists did not speak of genes as conveyors of "information" in such words, but if they had, they would not have hesitated to quantify the amount of information that genes need to convey as vast. The idea that information might reside in a chemical in the same way that it exists in text--as a finite alphabet of letters arranged in a sequence of unlimited length--had not yet been conceived. It would emerge upon the discovery of DNA's structure, but few researchers imagined that DNA's structure had much to say about genetics.
In the 1950s, only a few groups made it their goal to determine the structure of DNA. These included an American group led by Linus Pauling, and two groups in Britain. At Cambridge University, Crick and Watson were building physical models using metal rods and balls, in which they incorporated the known chemical structures of the nucleotides, as well as the known position of the linkages joining one nucleotide to the next along the polymer. At King's College, London, Maurice Wilkins and Rosalind Franklin were examining x-ray diffraction patterns of DNA fibers.
A key inspiration in the work of all of these teams was the discovery in 1948 by Pauling that many proteins included helical (see alpha helix) shapes. Pauling had deduced this structure from x-ray patterns. Even in the initial crude diffraction data from DNA, it was evident that the structure involved helices. But this insight was only a beginning. There remained the questions of how many strands came together, whether this number was the same for every helix, whether the bases pointed toward the helical axis or away, and ultimately what were the explicit angles and coordinates of all the bonds and atoms. Such questions motivated the modeling efforts of Watson and Crick.
In their modeling, Watson and Crick restricted themselves to what they saw as chemically and biologically reasonable. Still, the breadth of possibilities was very wide. A breakthrough occurred in 1952, when Erwin Chargaff visited Cambridge and inspired Crick with a description of experiments Chargaff had published in 1947. Chargaff had observed that the proportions of the four nucleotides vary between one DNA sample and the next, but that for particular pairs of nucleotides -- adenine and thymine, guanine and cytosine -- the two nucleotides are always present in equal proportions.
Watson and Crick had begun to contemplate double helical arrangements, and they saw that by reversing the directionality of one strand with respect to the other, they could provide an explanation for Chargaff's puzzling finding. This explanation was the complementary pairing of the bases, which also had the effect of ensuring that the distance between the phosphate chains did not vary along a sequence. Watson and Crick were able to discern that this distance was constant and to measure its exact value of 2 nanometers from an X-ray pattern obtained by Franklin. The same pattern also gave them the 3.4 nanometer-per-10 bp "pitch" of the helix. The pair quickly converged upon a model, which they announced before Franklin herself published any of her work.
The great assistance Watson and Crick derived from Franklin's data has become a subject of controversy, and it has angered people who believe Franklin has not received the credit due to her. The most controversial aspect is that Franklin's critical X-ray pattern was shown to Watson and Crick without Franklin's knowledge or permission. Wilkins showed it to them at his lab while Franklin was away.
Watson and Crick's model attracted great interest immediately upon its presentation. Arriving at their conclusion on February 21, 1953, Watson and Crick made their first announcement on February 28. Their paper 'A Structure for Deoxyribose Nucleic Acid' was published on April 25. In an influential presentation in 1957, Crick laid out the "Central Dogma", which foretold the relationship between DNA, RNA, and proteins, and articulated the "sequence hypothesis." A critical confirmation of the replication mechanism that was implied by the double-helical structure followed in 1958 in the form of the Meselson-Stahl experiment. Work by Crick and coworkers deciphered the genetic code not long afterward. These findings represent the birth of molecular biology.
Watson, Crick, and Wilkins were awarded a Nobel Prize in 1962, by which time Franklin had died.